 View full size
View full size
- Analiza kwasów nukleinowych
- Analiza białek
- Odczynniki biochemiczne
- Enzymy
- Edytowanie genów
- Klonowanie
- Diagnostyka kliniczna
- Human Identification STR kits
- Sprzęt laboratoryjny
- Oprogramowanie
- A&A Biotechnology
- AdvancedSeq
- BioDynami
- Plant Cell Technology
Aktualności
-
RANK 2026
Zapraszamy do odwiedzenia nas podczas jubileuszowej 20. edycji konferencji naukowej RANK 2026, która odbędzie się w dniach 18–19 marca w Pardubicach, w klubie ABC. Konferencja jest organizowana przez ...
Czytaj więcej -
XXXV. Izakovičov memoriál 2025
We are pleased to announce our participation in the prestigious XXXV. Izakovič Memorial 2025, which will take place on October 8–10, 2025 at the Grandhotel Praha, Tatranská Lomnica. The Izakovič Memo...
Czytaj więcej -
1st Czechoslovak Congress of Medical Genetics 2025
In the spring, we will participate in the 1st Czechoslovak Congress of Medical Genetics, which will take place from April 2–4, 2025, at the Cultural and Congress Center Elektra in the spa town of Luha...
Czytaj więcej
 View full size
View full size
Description:
The enzyme mix was developed to generate random fragmentation of genomic DNA with blunt ends in a single step. The fragmented DNA can be used for applications such as Next-Generation Sequencing (NGS), blunt end ligation and PCR cloning.
Compared to mechanical shearing methods, enzymatic DNA fragmentation is significantly easier and more efficient. It allows convenient handling of multiple samples simultaneously, minimizes sample loss, and eliminates the need for expensive equipment.
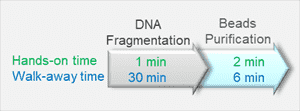
The illustration below shows the workflow for DNA fragmentation for NGS:
Step 1: DNA fragmentation – 1 minute hands-on, 30 minutes incubation
Step 2: Magnetic bead purification – 2 minutes hands-on, 6 minutes automated incubation
This streamlined process saves time, allowing most of the workflow to run unattended.
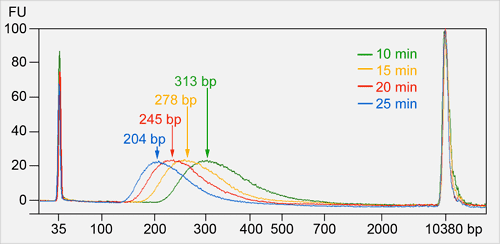
The graph demonstrates how DNA fragmentation progresses depending on incubation time using this enzymatic mix. Each curve represents a different incubation duration (10, 15, 20, 25 minutes) and the resulting average DNA fragment sizes (204–313 bp). The product allows precise control over fragment size – longer incubation yields smaller DNA fragments.
Features:
- Fast fragmentation: Efficient enzymatic DNA fragmentation in 30–45 minutes
- Ease of use: Only 3 minutes hands-on time – minimal manual steps
- Sample flexibility: Compatible with EDTA-free DNA and DNA in TE buffer
- No expensive equipment needed: No need for complex mechanical shearing systems
- NGS-optimized: Ideal for NGS library prep, PCR cloning, TA-ligation, and blunt-end ligation without detectable sequencing bias
- Fragment size control: Easily adjusted by changing incubation time – longer incubation results in shorter fragments
Product sizes:
50 reactions (Cat. No. 40063S)
200 reactions (Cat. No. 40063L)